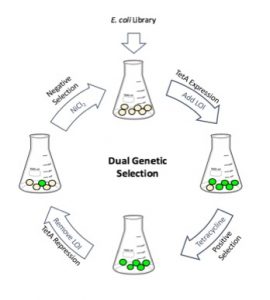
Our long-term research goal is to develop a general method to rapidly and easily monitor small molecules relevant to the study of biology and the environment. A research objective of the lab is to engineer riboswitch-based biosensors to monitor intracellular concentrations of small organic molecules. Riboswitches are naturally occurring non-coding RNA elements that act as direct sensors of diverse small molecules and can signal molecular recognition through altered protein expression. We have used a naturally occurring riboswitch for an important bacterial signaling molecule, cyclic diguanylate (c-di-GMP), to create a c-di-GMP sensor. When applied to the water-borne pathogen, Vibrio cholerae, the sensor was able to identify specific extracellular signals that affect intracellular c-di-GMP levels and likely contribute to the persistence of the bacteria in aquatic reservoirs. More recently, a novel riboswitch was engineered by the Liu Lab through in vivo directed evolution. This de novo development of a riboswitch is transformative as it provides a general platform from which many more synthetic riboswitches may be discovered, addressing a major obstacle to broadening the scope of riboswitch-based technology.